Lets write a Convolutional Neural Networks From Scratch. Writing Convoluitional Nerual Networks from Scratch is one of the challenging thing to be even for experienced person because we have been using frameworks like PyTorch to train and slowly forgetting basics of it.
What will you do when you are stuck in a village with no electricity for 4 days and you only have a pen and paper? For me, I wrote a Convolutional Neural Networks from Scratch on paper. Once again, high credit goes to the pandemic Corona Virus, without it, I would not have lived as a farmer once more and the idea of ' from scratch' arose.
I am sorry for not using a single image here on this blog because I was low on data and this entire blog is written on markdown(sometimes latex) only so the text format might seem a little disturbing also.
If you are here, then you are encouraged to look at the below 3 blog posts(serially) of mine(most of the concepts on this blog are taken from the below posts):
- Writing a Feedforward Neural Network from Scratch on Python
- This post gives a brief introduction to an OOP concept of making a simple Keras-like ML library.
- A gentle introduction to backpropagation and gradient descent from scratch.
- Writing top Machine Learning Optimizers from scratch on Python
- Gives introduction and python code to optimizers like
GradientDescentand ADAM`.
- Gives introduction and python code to optimizers like
- Writing a Image Processing Codes from Scratch on Python
- This post gives a brief introduction to convolution operation and RGB to grayscale conversion from scratch.
- We will be using the same convolution concept here on this blog.
Updates:
- 2020/06/05: Published blog.
- 2022/11/10: Fixed errors in the derivative.
1.1 What this Convolutional Neural Networks from Scratch blog will cover?
- Includes
Feed forwardlayer - A gentle introduction to
Conv2d - Includes
Dropoutlayer - Includes
Pool2dlayer - Includes
Flattenlayer - Test Cases with different architectures(4 of them) on
MNISTdataset - Bonus Topics
Testing a model will require huge time, my system is Dell I5 with 8GB RAM and 256GB SSD. And I tested these models on my local machine. It had taken nearly a week to find the test cases and improve the overall concepts. Sometimes, I had to sleep on my laptop for saving battery power so some epochs might be seen taking 4+hours of time. And yes, I used mobile data to post this blog.
2 Preliminary Concepts for Convolutional Neural Networks from Scratch
- Every layer will have the common methods(doing so will ease the overhead of method calling):
set_output_shapeapply_activationConv2dcan have functions likereluand convolution operation happens hereFFLwill use theactivation_fnmethod on a linear combination of input, weights, and biases.Pool2dwill perform pooling operations likemax, min, averageDropoutwill perform setting input to 0 randomlyFlattenwill convert feature vectores to 1d vector
backpropagateConv2dwill use the delta term of the next layer to find the delta term and delta parametersFFLPool2d: error is backpropagated from the index of the output of this layerDropout: propagate error through non-zero output unitsFlatten: propagate error of next layer to previous by reshaping to input shape
3 Steps
- Prepare layers
- Prepare stacking class
- Prepare Optimizers
3.1 Prepare Layers
Let's prepare layers from scratch for Convolutional Neural Networks from Scratch.
3.1.1 Feedforward Layer
For a typical Convolutional Neural Networks from Scratch, we need a feedforward layer as well. I am not going to explain much more here because a previous post about Writing a Feedforward Neural Network from Scratch on Python has explained already.
class FFL():
def __init__(self, input_shape=None, neurons=1, bias=None, weights=None, activation=None, is_bias = True):
np.random.seed(100)
self.input_shape = input_shape
self.neurons = neurons
self.isbias = is_bias
self.name = ""
self.w = weights
self.b = bias
if input_shape != None:
self.output_shape = neurons
if self.input_shape != None:
self.weights = weights if weights != None else np.random.randn(self.input_shape, neurons)
self.parameters = self.input_shape * self.neurons + self.neurons if self.isbias else 0
if(is_bias):
self.biases = bias if bias != None else np.random.randn(neurons)
else:
self.biases = 0
self.out = None
self.input = None
self.error = None
self.delta = None
activations = ["relu", "sigmoid", "tanh", "softmax"]
self.delta_weights = 0
self.delta_biases = 0
self.pdelta_weights = 0
self.pdelta_biases = 0
if activation not in activations and activation != None:
raise ValueError(f"Activation function not recognised. Use one of {activations} instead.")
else:
self.activation = activation
def activation_dfn(self, r):
"""
A method of FFL to find the derivative of a given activation function.
"""
if self.activation is None:
return np.ones(r.shape)
if self.activation == 'tanh':
return 1 - r ** 2
if self.activation == 'sigmoid':
# r = self.activation_fn(r)
return r * (1 - r)
if self.activation == "softmax":
soft = self.activation_fn(r)
diag_soft = soft*(1- soft)
return diag_soft
if self.activation == 'relu':
r[r < 0] = 0
r[r>=1]=1
return r
return r
def activation_fn(self, r):
"""
A method of FFL that contains the operation and definition of a given activation function.
"""
if self.activation == 'relu':
r[r < 0] = 0
return r
if self.activation == None or self.activation == "linear":
return r
if self.activation == 'tanh':
return np.tanh(r)
if self.activation == 'sigmoid':
return 1 / (1 + np.exp(-r))
if self.activation == "softmax":
r = r - np.max(r)
s = np.exp(r)
return s / np.sum(s)
def apply_activation(self, x):
soma = np.dot(x, self.weights) + self.biases
self.out = self.activation_fn(soma)
return self.out
def set_n_input(self):
self.weights = self.w if self.w != None else np.random.normal(size=(self.input_shape, self.neurons))
def backpropagate(self, nx_layer):
self.error = np.dot(nx_layer.weights, nx_layer.delta)
self.delta = self.error * self.activation_dfn(self.out)
self.delta_weights += self.delta * np.atleast_2d(self.input).T
self.delta_biases += self.delta
def set_output_shape(self):
self.set_n_input()
self.output_shape = self.neurons
self.get_parameters()
def get_parameters(self):
self.parameters = self.input_shape * self.neurons + self.neurons if self.isbias else 0
return self.parameters 3.1.2 Conv2d Layer
This layer will be the crucial layer for Convolutional Neural Networks from Scratch.
3.1.2.1 Let's initialize it first.
class Conv2d():
def __init__(self, input_shape=None, filters=1, kernel_size = (3, 3), isbias=True, activation=None, stride=(1, 1), padding="zero", kernel=None, bias=None):
self.input_shape = input_shape
self.filters = filters
self.isbias = isbias
self.activation = activation
self.stride = stride
self.padding = padding
self.p = 1 if padding != None else 0
self.bias = bias
self.kernel = kernel
if input_shape != None:
self.kernel_size = (kernel_size[0], kernel_size[1], input_shape[2], filters)
self.output_shape = (int((input_shape[0] - kernel_size[0] + 2 * self.p) / stride[0]) + 1,
int((input_shape[1] - kernel_size[1] + 2 * self.p) / stride[1]) + 1, filters)
self.set_variables()
self.out = np.zeros(self.output_shape)
else:
self.kernel_size = (kernel_size[0], kernel_size[1]) Initializing takes:-
input_shape:- It is the input shape of this layer. It will include a tuple of(rows, cols, num_channels). For any noninput layer, it will default i.e.None.filters:- How many kernels or filters are we using?kernel_size:- It is the size of convoluting tuple of matrix or filter's(row, cols). Later we will create a kernel of shaperows, cols, input_channels, num_filters.isbias: Boolean value for whether we will use bias or not.activaiton: Activation function.tride: A tuple indicating a step of convolution operation per row, column.padding: String indicating what operation will be done on borders, available among[zeros, same].kernel: A convoluting matrix. Recommended not to use it.bias: A array of shape(num_filters, 1)will be added after each convolution operation.
A few important things inside this method are:-
-
The
output_shapeof any convolution layer will be:
\begin{equation}
W = \frac{(w-f+2*p)}{s} + 1
\end{equation}Where W is output width or shape and w is input width or shape.\
f is filter width.\
p is padding(1 if used)\
s is stride width or shape\ - The method
set_variables()sets all the important parameters needed for training. self.outwill be the output of this layer andself.doutwill be the delta out.self.deltawill be the delta term of this layer
3.1.2.2 set_variable() method
def set_variables(self):
self.weights = self.init_param(self.kernel_size)
self.biases = self.init_param((self.filters, 1))
self.parameters = np.multiply.reduce(self.kernel_size) + self.filters if self.isbias else 1
self.delta_weights = np.zeros(self.kernel_size)
self.delta_biases = np.zeros(self.biases.shape) - To make our optimization easier, we are naming filters as weights.
- The method
init_param()initializes the parameter from the random normal sample.
def init_param(self, size):
stddev = 1/np.sqrt(np.prod(size))
return np.random.normal(loc=0, scale=stddev, size=size)3.1.2.3 Prepare Activation Functions
def activation_fn(self, r):
"""
A method of FFL that contains the operation and definition of a given activation function.
"""
if self.activation == None or self.activation == "linear":
return r
if self.activation == 'tanh': #tanh
return np.tanh(r)
if self.activation == 'sigmoid': # sigmoid
return 1 / (1 + np.exp(-r))
if self.activation == "softmax":# stable softmax
r = r - np.max(r)
s = np.exp(r)
return s / np.sum(s)
Recall the mathematics,
\begin{equation}
i. tanh(soma) = \frac{1-soma}{1+soma}
\end{equation}
\begin{equation}
ii. linear(soma) = soma
\end{equation}
\begin{equation}
iii. sigmoid(soma) = \frac{1}{1 + exp^{(-soma)}}
\end{equation}
\begin{equation}
iv. relu(soma) = \max(0, soma)
\end{equation}
\begin{equation}
v. softmax(x_j) = \frac{exp^{(xj)}}{\sum{i=1}^n{exp^{(x_i)}}}
\end{equation}
\begin{equation}
Where, soma = XW + \theta
\end{equation}
And W is the weight vector of shape (n, w). X is the input vector of shape (m, n) and 𝜃 is the bias term of shape w, 1.
3.1.2.4 Prepare derivative of Activation Function
def activation_dfn(self, r):
"""
A method of FFL to find the derivative of a given activation function.
"""
if self.activation is None:
return np.ones(r.shape)
if self.activation == 'tanh':
return 1 - r ** 2
if self.activation == 'sigmoid':
return r * (1 - r)
if self.activtion == 'softmax':
soft = self.activation_fn(r)
return soft * (1 - soft)
if self.activation == 'relu':
r[r<0] = 0
r[>=1]=1
return rLet's revise a bit of calculus.
Why do we need derivative?
While doing Convolutional Neural Networks from Scratch, we need to do few derivatives.
Well, if you are here then you already know that gradient descent is based on the derivatives(gradients) of activation functions and errors. So we need to perform this derivative. But you are on your own to perform calculations. I will also explain the gradient descent later.
\begin{equation}
i. \frac{d(linear(x))}{d(x)} = 1
\end{equation}
\begin{equation}
ii. \frac{d(sigmoid(x))}{d(x)} = sigmoid(x)(1- sigmoid(x))
\end{equation}
\begin{equation}
iii. \frac{d(tanh(x))}{d(x)} = 1-tanh(x)**2
\end{equation}
\begin{equation}
iv. \frac{d(relu(x))}{d(x)} = 1 if x>=1 else 0
\end{equation}
\begin{equation}
v. \frac{d(softmax(x_j))}{d(x_k)} = softmax(x_j)(1- softmax(x_j)) \space when \space j = k \space else\
\space -softmax({x_j}).softmax({x_k})
\end{equation}
For the sake of simplicity, we use the case of j = k for softmax.
3.1.2.5 Prepare a method to do feedforward on this layer
def apply_activation(self, image):
for f in range(self.filters):
image = self.input
kshape = self.kernel_size
if kshape[0] % 2 != 1 or kshape[1] % 2 != 1:
raise ValueError("Please provide odd length of 2d kernel.")
if type(self.stride) == int:
stride = (stride, stride)
else:
stride = self.stride
shape = image.shape
if self.padding == "zero":
zeros_h = np.zeros((shape[1], shape[2])).reshape(-1, shape[1], shape[2])
zeros_v = np.zeros((shape[0]+2, shape[2])).reshape(shape[0]+2, -1, shape[2])
padded_img = np.vstack((zeros_h, image, zeros_h)) # add rows
padded_img = np.hstack((zeros_v, padded_img, zeros_v)) # add cols
image = padded_img
shape = image.shape
elif self.padding == "same":
h1 = image[0].reshape(-1, shape[1], shape[2])
h2 = image[-1].reshape(-1, shape[1], shape[2])
padded_img = np.vstack((h1, image, h2)) # add rows
v1 = padded_img[:, 0].reshape(padded_img.shape[0], -1, shape[2])
v2 = padded_img[:, -1].reshape(padded_img.shape[0], -1, shape[2])
padded_img = np.hstack((v1, padded_img, v2)) # add cols
image = padded_img
shape = image.shape
elif self.padding == None:
pass
rv = 0
cimg = []
for r in range(kshape[0], shape[0]+1, stride[0]):
cv = 0
for c in range(kshape[1], shape[1]+1, stride[1]):
chunk = image[rv:r, cv:c]
soma = (np.multiply(chunk, self.weights[:, :, :, f]))
summa = soma.sum()+self.biases[f]
cimg.append(summa)
cv+=stride[1]
rv+=stride[0]
cimg = np.array(cimg).reshape(int(rv/stride[0]), int(cv/stride[1]))
self.out[:, :, f] = cimg
self.out = self.activation_fn(self.out)
return self.outI have linked a post about convolution operation at the top of this blog. The only important part here is:-
- For each filter
- do elementwise matrix multiplication and sum them all(of each channels also)
- Then add bias term
- Output of this filter will have channel(not a real color channel) of
num_filters
- Finally apply the activation function on this output.
It is clear that, if a layer has 5 filters then the output of this layer will have 5 channels also.
3.1.2.6 Prepare Method for Backpropagation
def backpropagate(self, nx_layer):
layer = self
layer.delta = np.zeros((layer.input_shape[0], layer.input_shape[1], layer.input_shape[2]))
image = layer.input
for f in range(layer.filters):
kshape = layer.kernel_size
shape = layer.input_shape
stride = layer.stride
rv = 0
i = 0
for r in range(kshape[0], shape[0]+1, stride[0]):
cv = 0
j = 0
for c in range(kshape[1], shape[1]+1, stride[1]):
chunk = image[rv:r, cv:c]
layer.delta_weights[:, :, :, f] += chunk * nx_layer.delta[i, j, f]
layer.delta[rv:r, cv:c, :] += nx_layer.delta[i, j, f] * layer.weights[:, :, :, f]
j+=1
cv+=stride[1]
rv+=stride[0]
i+=1
layer.delta_biases[f] = np.sum(nx_layer.delta[:, :, f])
layer.delta = layer.activation_dfn(layer.delta)Backpropagating error from the Convolution layer is a really hard and challenging task. I have tried my best to do the right way of backpropagation but I still have doubt about it. Some really awesome articles like below can help to understand these things:-
- Convolutional Neural Network from Ground Up
- A Gentle Introduction to CNN
- Training a Convolutional Neural Networks from Scratch
For understanding how to pass errors and find the delta terms for parameters:
- The delta term for this layer will be equal to the shape of the input i.e.
(input_row, input_cols, input_channels). - We will also take the input to this layer into consideration.
- For each filter:-
- Loop through each row and col just like the convolution operation
- Get the chunk or part of the image and multiply it with the delta term of the next layer to get the delta filter(weight)
- i.e.
layer.delta_weights[:, :, :, f] += chunk * nx_layer.delta[i, j, f]a trick to understanding the delta of the next layer is by revisiting the input and output shape of the layer. For a layer with 5 filters, the output will have 5 channels. And the delta term of the next layer will have the same number of channels. Hence we are giving[i, j, f]. Note that for every step on the input image(i.e step on row and col),i,jwill increase by 1. Initially,layer.delta_weights[:, :, :, f]will be all 0s but it will change by visiting every chunk. Since we have a filter of shape(row, col, channels, num_filters), delta_weights is updated for each filter by adding it with the multiplication of each chunk with the corresponding next layer's delta. - Delta term of this layer will have shape of
(input_rows, input_cols, channels)i.e equal to input shape. Hence we will set the delta term using the number of channels on this layer's filters. We will add the delta term for that chunk using each filter. Because each filter is responsible for the error and the contribution of each filter must be taken equally. Thelayer.delta[rv:r, cv:c, :] += nx_layer.delta[i, j, f] * layer.weights[:, :, :, f]is here to do this task. - We increase I after completing the row and j after completing the column.
iandjare used to getting values from the delta of the next layer.
- i.e.
- We sum the delta term of this filter to get
delta_biasesdue to this filter.
- Finally, we get the delta of this layer by applying the derivative of the activation function of this layer.
There are different approaches than this one of doing backpropagation on the Convolution layer. I found this one to be working on my case(i wrote this approach). Please try to visit one of the above links for more explanation.
Please test your class like below:-
img = xt[0]
conv = Conv2d()
conv.input=img
conv.weights = np.array([[1, 0, -1], [1, 0, -1], [1, 0, -1]]).reshape(3, 3, 1, 1)
conv.biases = np.zeros(1)
conv.out = np.zeros((28, 28, 1))
cout = conv.apply_activation(img)
plt.imshow(cout.reshape(28, 28)) Where xt is an image array of shape (28, 28, 1) from mnist.
3.1.3 Dropout Layer
The main concept behind the dropout layer is to forget some of the inputs to the current layer forcefully. Doing so will reduce the risk of overfitting the model.
class Dropout:
def __init__(self, prob = 0.5):
self.input_shape=None
self.output_shape = None
self.input_data= None
self.output = None
self.isbias = False
self.activation = None
self.parameters = 0
self.delta = 0
self.weights = 0
self.bias = 0
self.prob = prob
self.delta_weights = 0
self.delta_biases = 0
def set_output_shape(self):
self.output_shape = self.input_shape
self.weights = 0
def apply_activation(self, x, train=True):
if train:
self.input_data = x
flat = np.array(self.input_data).flatten()
random_indices = np.random.randint(0, len(flat), int(self.prob * len(flat)))
flat[random_indices] = 0
self.output = flat.reshape(x.shape)
return self.output
else:
self.input_data = x
self.output = x / self.prob
return self.output
def activation_dfn(self, x):
return x
def backpropagate(self, nx_layer):
if type(nx_layer).__name__ != "Conv2d":
self.error = np.dot(nx_layer.weights, nx_layer.delta)
self.delta = self.error * self.activation_dfn(self.out)
else:
self.delta = nx_layer.delta
self.delta[self.output == 0] = 0- Some of the parameters like
weights,biasesare actually not available on the Dropout layer but I am using this for the sake of simplicity while working with a stack of layers. - The input shape and output shape of the Dropout layer will be the same, what differs is the value. Where some will be set to 0 i.e forgotten randomly.
- The method
apply_activationperforms the dropout operation.- The easier way is to first convert it to a 1d vector(by NumPy's
flatten) and take random indices from a given probability. - Then we set the element of those random indices to 0 and return the reshaped new array as the output of this layer.
- The easier way is to first convert it to a 1d vector(by NumPy's
- The method
backpropagateperforms the backpropagation operation on error.- We set the delta to
0if the recent output of this layer is 0, else leave it as it is.
- We set the delta to
- Note:- In the testing phase, forward propagation will be different. Entire activation is reduced by a factor. So we are also giving a training parameter to
apply_activation.
Lets test our class:-
x = np.arange(0, 100).reshape(10, 10)
dp = Dropout()
dp.apply_activation(x)3.1.4 Pooling Layer
A convolutional neural network's work can be thought of as:
- Take an image where we want to perform a convolution.
- Take a lens(will be filtered) and place it over an image.
- Slide the lens over an image and find the important features.
- We find features using different lenses.
- Once we found certain features under our boundary, we pass those feature maps to the next scanning place or we can do pooling.
- Pooling can be thought of as zooming out, or we make the remaining image a little smaller, by this way more important features will be seen. Or in another way, scan from a bit far and take only the important parts.
A pooling operation works in a similar way to convolution but instead of matrix multiplication, we do a different operation. The output of a pooling layer will be:-
\begin{equation}
w = \frac{W-f + 2p}{s} + 1
\end{equation}
where w is the new width, W is the old or input width, f is kernel width, p is padding. I am not using padding right now for the operation.
3.1.4.1 Initializing a Class
class Pool2d:
def __init__(self, kernel_size = (2, 2), stride=None, kind="max", padding=None):
self.input_shape=None
self.output_shape = None
self.input_data= None
self.output = None
self.isbias = False
self.activation = None
self.parameters = 0
self.delta = 0
self.weights = 0
self.bias = 0
self.delta_weights = 0
self.delta_biases = 0
self.padding = padding
self.p = 1 if padding != None else 0
self.kernel_size = kernel_size
if type(stride) == int:
stride = (stride, stride)
self.stride = stride
if self.stride == None:
self.stride = self.kernel_size
self.pools = ['max', "average", 'min']
if kind not in self.pools:
raise ValueError("Pool kind not understood.")
self.kind = kindMost of the attributes are common to the `Convolution layer.
- Just like Keras, we will set the
stridetokernel_sizeif nothing is given. - The pool is a list of available pooling types. Currently, I have only included 3.
3.1.4.2 Method set_output_shape
As always, this method will always be called from the stacking class.
def set_output_shape(self):
self.output_shape = (int((self.input_shape[0] - self.kernel_size[0] + 2 * self.p) / self.stride[0] + 1),
int((self.input_shape[1] - self.kernel_size[1] + 2 * self.p) / self.stride[1] + 1), self.input_shape[2]) 3.1.4.3 Feedforward or apply_activation method
This method will perform the real pooling operation indicated above.
def apply_activation(self, image):
stride = self.stride
kshape = self.kernel_size
shape = image.shape
self.input_shape = shape
self.set_output_shape()
self.out = np.zeros((self.output_shape))
for nc in range(shape[2]):
cimg = []
rv = 0
for r in range(kshape[0], shape[0]+1, stride[0]):
cv = 0
for c in range(kshape[1], shape[1]+1, stride[1]):
chunk = image[rv:r, cv:c, nc]
if len(chunk) > 0:
if self.kind == "max":
chunk = np.max(chunk)
if self.kind == "min":
chunk = np.min(chunk)
if self.kind == "average":
chunk = np.mean(chunk)
cimg.append(chunk)
else:
cv-=cstep
cv+=stride[1]
rv+=stride[0]
cimg = np.array(cimg).reshape(int(rv/stride[0]), int(cv/stride[1]))
self.out[:,:,nc] = cimg
return self.outLets take an example:-
$$
\begin{equation}
x =
\begin{pmatrix}
1 & 2 & 3 & 1 \\
11 & 12 & 4 & 10 \\
101 & 11 & 88 & 10 \\
10 & 11 & 11 & 5 \end{pmatrix}
\end{equation}
$$
After maxpool of size (2, 2) and stride (2, 2):-
- First our pointer will be 0 for row/col i.e
curr_pointer = (0, 0)and window will be values ofcurr_pointer:curr_pointer+kernel_size-1. - In other words, our first window will be
[[1 2] [11, 12]]. - Then for the max pool, the maximum value on this window is 12, so 12 is taken, if the average pool then the output of this window will be
6.5i.e average of1, 2, 11, 12. - Then current pointer of row will be
prev_pointer[0]+stride[0] - Now the new window will be
[[3 1] [4 10]]and the max pool will be10. - Now we have reached the end of this row, we will increase the column. Then the current pointer will be
curr_pointer + (0, stride[1]-1).
Maxpooling of 𝑥:
$$
\begin{pmatrix}
12 & 10 \\
101 & 88 \end{pmatrix}
$$
In a simpler way, we took only those values which contribute to high value.
3.1.4.4 Backpropagate Method
def backpropagate(self, nx_layer):
"""
Gradients are passed through an index of the latest output value.
"""
layer = self
stride = layer.stride
kshape = layer.kernel_size
image = layer.input
shape = image.shape
layer.delta = np.zeros(shape)
cimg = []
rstep = stride[0]
cstep = stride[1]
for f in range(shape[2]):
i = 0
rv = 0
for r in range(kshape[0], shape[0]+1, rstep):
cv = 0
j = 0
for c in range(kshape[1], shape[1]+1, cstep):
chunk = image[rv:r, cv:c, f]
dout = nx_layer.delta[i, j, f]
if layer.kind == "max":
p = np.max(chunk)
index = np.argwhere(chunk == p)[0]
layer.delta[rv+index[0], cv+index[1], f] = dout
if layer.kind == "min":
p = np.min(chunk)
index = np.argwhere(chunk == p)[0]
layer.delta[rv+index[0], cv+index[1], f] = dout
if layer.kind == "average":
p = np.mean(chunk)
layer.delta[rv:r, cv:c, f] = dout
j+=1
cv+=cstep
rv+=rstep
i+=1The main idea behind the backpropagation on Pooling Layer is:-
- If pooling is
Maxthen an error is passed through an index of the largest value on the chunk. - If pooling is
Minthen error is passed through an index of the smallest value on the chunk. - If pooling is
averagethen an error is passed through entire indices on a chunk
Since the output shape and input shape's number of the channel remain the same, we loop through each channel and get the delta for each channel. So we are not adding the delta term.
Lets test our pooling class:
pool = Pool2d(kernel_size=(7, 7), kind="max")
test = np.random.randint(1, 100, (32, 32, 3))
o = pool.apply_activation(test)If you don't get any errors then, great let's proceed. Else please see the reference file on GitHub.
3.1.5 Flatten Layer
Flatten layer's main task is to take entire feature maps of the previous layer and make a 1d vector from it. Flatten layer is used before passing a result of convolution to classification layers.
Let the input to Flatten be (3, 3, 3).
$$
\begin{equation}
x =
\begin{pmatrix}
\begin{pmatrix}
1 & 2 & 3\end{pmatrix}
\begin{pmatrix}
1 & 11 & 12\end{pmatrix}
\begin{pmatrix}
4 & 10 & 1\end{pmatrix}\\
\begin{pmatrix}
101 & 11 & 88\end{pmatrix}
\begin{pmatrix}
10 & 11 & 11\end{pmatrix}
\begin{pmatrix}
5 & 111 & 33\end{pmatrix}\\
\begin{pmatrix}
9 & 11 & 123\end{pmatrix}
\begin{pmatrix}
66 & 110 & 12\end{pmatrix}
\begin{pmatrix}
100 & 11 & 12\end{pmatrix}
\end{pmatrix}
\end{equation}
$$
Flatten output will be:
$$
\begin{equation}
\begin{pmatrix}
1 & 2 & 3&
1 & 11 & 12&
4 & 10 & 1&
101 & 11 & 88&
10 & 11 & 11&
5 & 111 & 33&
9 & 11 & 123&
66 & 110 & 12&
100 & 11 & 12&
\end{pmatrix}
\end{equation}
$$
class Flatten:
def __init__(self, input_shape=None):
self.input_shape=None
self.output_shape = None
self.input_data= None
self.output = None
self.isbias = False
self.activation = None
self.parameters = 0
self.delta = 0
self.weights = 0
self.bias = 0
self.delta_weights = 0
self.delta_biases = 0
def set_output_shape(self):
self.output_shape = (self.input_shape[0] * self.input_shape[1] * self.input_shape[2])
self.weights = 0
def apply_activation(self, x):
self.input_data = x
self.output = np.array(self.input_data).flatten()
return self.output
def activation_dfn(self, x):
return x
def backpropagate(self, nx_layer):
self.error = np.dot(nx_layer.weights, nx_layer.delta)
self.delta = self.error * self.activation_dfn(self.out)
self.delta = self.delta.reshape(self.input_shape)Note: There will be no attributes like weights, biases on Flatten layer but I used to make it work on doing optimization
- The output shape of this layer will be the multiplication of
(num_rows, num_cols, num_channels). - Since this layer will be connected before the feedforward layer, error and delta terms are calculated like on the feedforward layer.
- The shape of the delta of this layer will be the shape of the input.
Lets test our flatten class:
x = np.array([[1, 1, 1], [1, 0, 1], [0, 1, 1], [0, 0, 1]])
f = Flatten()
print(f.apply_activation(test)) If you got output like the below, then cool:-
[1, 1, 1, 1, 0, 1, 0, 1, 1, 0, 0, 1]
3.2 Creating a Stacking class
We will name it CNN.
As previous feedforward post, this will perform all the tasks like training, testing and so on.
3.2.1 Initializing a class
Please refer to the previous post about Feedforward Neural Networks for more explanation.
class CNN():
def __init__(self):
self.layers = []
self.info_df = {}
self.column = ["LName", "Input Shape", "Output Shape", "Activation", "Bias"]
self.parameters = []
self.optimizer = ""
self.loss = "mse"
self.lr = 0.01
self.mr = 0.0001
self.metrics = []
self.av_optimizers = ["sgd", "momentum", "adam"]
self.av_metrics = ["mse", "accuracy", "cse"]
self.av_loss = ["mse", "cse"]
self.iscompiled = False
self.model_dict = None
self.out = []
self.eps = 1e-15
self.train_loss = {}
self.val_loss = {}
self.train_acc = {}
self.val_acc = {} 3.2.2 Creating a add Method
Please refer to the previous post for more explanation.
def add(self, layer):
if(len(self.layers) > 0):
prev_layer = self.layers[-1]
if prev_layer.name != "Input Layer":
prev_layer.name = f"{type(prev_layer).__name__}{len(self.layers) - 1}"
if layer.input_shape == None:
if type(layer).__name__ == "Flatten":
ops = prev_layer.output_shape[:]
if type(prev_layer).__name__ == "Pool2d":
ops = prev_layer.output_shape[:]
elif type(layer).__name__ == "Conv2d":
ops = prev_layer.output_shape[:]
if type(prev_layer).__name__ == "Pool2d":
ops = prev_layer.output_shape
elif type(layer).__name__ == "Pool2d":
ops = prev_layer.output_shape[:]
if type(prev_layer).__name__ == "Pool2d":
ops = prev_layer.output_shape[:]
else:
ops = prev_layer.output_shape
layer.input_shape = ops
layer.set_output_shape()
layer.name = f"Out Layer({type(layer).__name__})"
else:
layer.name = "Input Layer"
if type(layer).__name__ == "Conv2d":
if(layer.output_shape[0] <= 0 or layer.output_shape[1] <= 0):
raise ValueError(f"The output shape became invalid [i.e. {layer.output_shape}]. Reduce filter size or increase image size.")
self.layers.append(layer)
self.parameters.append(layer.parameters)3.2.3 Writing a summary method:
Please refer to the previous post for more explanation.
def summary(self):
lname = []
linput = []
loutput = []
lactivation = []
lisbias = []
lparam = []
for layer in self.layers:
lname.append(layer.name)
linput.append(layer.input_shape)
loutput.append(layer.output_shape)
lactivation.append(layer.activation)
lisbias.append(layer.isbias)
lparam.append(layer.parameters)
model_dict = {"Layer Name": lname, "Input": linput, "Output Shape": loutput,
"Activation": lactivation, "Bias": lisbias, "Parameters": lparam}
model_df = pd.DataFrame(model_dict).set_index("Layer Name")
print(model_df)
print(f"Total Parameters: {sum(lparam)}")Test the class:
m = CNN()
m.add(Conv2d(input_shape = (28, 28, 1), filters = 2, padding=None, kernel_size=(3, 3), activation="relu"))
m.add(Conv2d(filters=4, kernel_size=(3, 3), padding=None, activation="relu"))
m.add(Pool2d(kernel_size=(2, 2)))
m.add(Conv2d(filters=6, kernel_size=(3, 3), padding=None, activation="relu"))
m.add(Conv2d(filters=8, kernel_size=(3, 3), padding=None, activation="relu"))
m.add(Pool2d(kernel_size=(2, 2)))
m.add(Dropout(0.1))
m.add(Flatten())
m.summary()3.2.4 Writing a train method
This method is identical to the train method of feeding a Forward Neural Network. Please refer to the previous post.
def train(self, X, Y, epochs, show_every=1, batch_size = 32, shuffle=True, val_split=0.1, val_x=None, val_y=None):
self.check_trainnable(X, Y)
self.batch_size = batch_size
t1 = time.time()
curr_ind = np.arange(0, len(X), dtype=np.int32)
if shuffle:
np.random.shuffle(curr_ind)
if type(val_x) != type(None) and type(val_y) != type(None):
self.check_trainnable(val_x, val_y)
print("\nValidation data found.\n")
else:
val_ex = int(len(X) * val_split)
val_exs = []
while len(val_exs) != val_ex:
rand_ind = np.random.randint(0, len(X))
if rand_ind not in val_exs:
val_exs.append(rand_ind)
val_ex = np.array(val_exs)
val_x, val_y = X[val_ex], Y[val_ex]
curr_ind = np.array([v for v in curr_ind if v not in val_ex])
print(f"\nTotal {len(X)} samples.\nTraining samples: {len(curr_ind)} Validation samples: {len(val_x)}.")
out_activation = self.layers[-1].activation
batches = []
len_batch = int(len(curr_ind)/batch_size)
if len(curr_ind)%batch_size != 0:
len_batch+=1
batches = np.array_split(curr_ind, len_batch)
print(f"Total {len_batch} batches, most batch has {batch_size} samples.\n")
for e in range(epochs):
err = []
for batch in batches:
a = []
curr_x, curr_y = X[batch], Y[batch]
b = 0
batch_loss = 0
for x, y in zip(curr_x, curr_y):
out = self.feedforward(x)
loss, error = self.apply_loss(y, out)
batch_loss += loss
err.append(error)
update = False
if b == batch_size-1:
update = True
loss = batch_loss/batch_size
self.backpropagate(loss, update)
b+=1
if e % show_every == 0:
train_out = self.predict(X[curr_ind])
train_loss, train_error = self.apply_loss(Y[curr_ind], train_out)
val_out = self.predict(val_x)
val_loss, val_error = self.apply_loss(val_y, val_out)
if out_activation == "softmax":
train_acc = train_out.argmax(axis=1) == Y[curr_ind].argmax(axis=1)
val_acc = val_out.argmax(axis=1) == val_y.argmax(axis=1)
elif out_activation == "sigmoid":
train_acc = train_out > 0.7
val_acc = val_out > 0.7
elif out_activation == None:
train_acc = abs(Y[curr_ind]-train_out) < 0.000001
val_acc = abs(Y[val_ex]-val_out) < 0.000001
self.train_loss[e] = round(train_error.mean(), 4)
self.train_acc[e] = round(train_acc.mean() * 100, 4)
self.val_loss[e] = round(val_error.mean(), 4)
self.val_acc[e] = round(val_acc.mean()*100, 4)
print(f"Epoch: {e}:")
print(f"Time: {round(time.time() - t1, 3)}sec")
print(f"Train Loss: {round(train_error.mean(), 4)} Train Accuracy: {round(train_acc.mean() * 100, 4)}%")
print(f'Val Loss: {(round(val_error.mean(), 4))} Val Accuracy: {round(val_acc.mean() * 100, 4)}% \n')
t1 = time.time()3.2.5 check_trainnable method
This method does the same work as the previous post's method.
def check_trainnable(self, X, Y):
if self.iscompiled == False:
raise ValueError("Model is not compiled.")
if len(X) != len(Y):
raise ValueError("Length of training input and label is not equal.")
if X[0].shape != self.layers[0].input_shape:
layer = self.layers[0]
raise ValueError(f"'{layer.name}' expects input of {layer.input_shape} while {X[0].shape[0]} is given.")
if Y.shape[-1] != self.layers[-1].neurons:
op_layer = self.layers[-1]
raise ValueError(f"'{op_layer.name}' expects input of {op_layer.neurons} while {Y.shape[-1]} is given.")3.2.6 Writing compiling method
This method is identical to the previous post's method.
def compile_model(self, lr=0.01, mr = 0.001, opt = "sgd", loss = "mse", metrics=['mse']):
if opt not in self.av_optimizers:
raise ValueError(f"Optimizer is not understood, use one of {self.av_optimizers}.")
for m in metrics:
if m not in self.av_metrics:
raise ValueError(f"Metrics is not understood, use one of {self.av_metrics}.")
if loss not in self.av_loss:
raise ValueError(f"Loss function is not understood, use one of {self.av_loss}.")
self.optimizer = opt
self.loss = loss
self.lr = lr
self.mr = mr
self.metrics = metrics
self.iscompiled = True
self.optimizer = Optimizer(layers=self.layers, name=opt, learning_rate=lr, mr=mr)
self.optimizer = self.optimizer.opt_dict[opt] In order to run properly, we need to have the Optimizer class defined. Please see this article about it.
3.2.7 Writing the feedforward method
This method is also the same as the previous post's method.
def feedforward(self, x, train=True):
if train:
for l in self.layers:
l.input = x
x = np.nan_to_num(l.apply_activation(x))
l.out = x
return x
else:
for l in self.layers:
l.input = x
if type(l).__name__ == "Dropout":
x = np.nan_to_num(l.apply_activation(x, train=train))
else:
x = np.nan_to_num(l.apply_activation(x))
l.out = x
return x3.2.8 Writing apply_loss method
This method is identical to the previous post's method.
def apply_loss(self, y, out):
if self.loss == "mse":
loss = y - out
mse = np.mean(np.square(loss))
return loss, mse
if self.loss == 'cse':
""" Requires out to be probability values. """
if len(out) == len(y) == 1: #print("Using Binary CSE.")
cse = -(y * np.log(out) + (1 - y) * np.log(1 - out))
loss = -(y / out - (1 - y) / (1 - out))
else: #print("Using Categorical CSE.")
if self.layers[-1].activation == "softmax":
"""if o/p layer's fxn is softmax then loss is y - out
check the derivation of softmax and cross-entropy with derivative"""
loss = y - out
loss = loss / self.layers[-1].activation_dfn(out)
else:
y = np.float64(y)
out += self.eps
loss = -(np.nan_to_num(y / out) - np.nan_to_num((1 - y) / (1 - out)))
cse = -np.sum((y * np.nan_to_num(np.log(out)) + (1 - y) * np.nan_to_num(np.log(1 - out))))
return loss, cse3.2.9 Writing the backpropagate method
This method is identical to the previous post's method.
def backpropagate(self, loss, update):
for i in reversed(range(len(self.layers))):
layer = self.layers[i]
if layer == self.layers[-1]:
if (type(layer).__name__ == "FFL"):
layer.error = loss
layer.delta = layer.error * layer.activation_dfn(layer.out)
layer.delta_weights += layer.delta * np.atleast_2d(layer.input).T
layer.delta_biases += layer.delta
else:
nx_layer = self.layers[i+1]
layer.backpropagate(nx_layer)
if update:
layer.delta_weights /= self.batch_size
layer.delta_biases /= self.batch_size
if update:
self.optimizer(self.layers)
self.zerograd()3.2.10zero_grad method
Same as previous.
def zerograd(self):
for l in self.layers:
try:
l.delta_weights=np.zeros(l.delta_weights.shape)
l.delta_biases = np.zeros(l.delta_biases.shape)
except:
pass3.2.11 predict method
Same as previous.
def predict(self, X):
out = []
if X.shape != self.layers[0].input_shape:
for x in X:
out.append(self.feedforward(x, train=False))
else:
out.append(self.feedforward(X, train = False))
return np.array(out)3.3 Preparing Optimizers
4 Testing with our Model
We just created Convolutional Neural Networks from Scratch but its time for a test.
4.1 Prepare datasets
Note:- More the training samples, more the performance of model(but not always). But more samples take more time to complete the epoch.
from keras.datasets import mnist
(x_train, y_train), (x_test, y_test) = mnist.load_data()
x = x_train.reshape(-1, 28 * 28)
x = (x-x.mean(axis=1).reshape(-1, 1))/x.std(axis=1).reshape(-1, 1)
x = x.reshape(-1, 28, 28, 1)
y = pd.get_dummies(y_train).to_numpy()
xt = x_test.reshape(-1, 28 * 28)
xt = (xt-xt.mean(axis=1).reshape(-1, 1))/xt.std(axis=1).reshape(-1, 1)
xt = xt.reshape(-1, 28, 28, 1)
yt = pd.get_dummies(y_test).to_numpy()4.2 Test 1:- Model with only one Conv2d and Output layer
m = CNN()
m.add(Conv2d(input_shape = (28, 28, 1), filters = 8, padding=None, kernel_size=(3, 3), activation="relu"))
m.add(Flatten())
m.add(FFL(neurons = 10, activation='softmax'))
m.compile_model(lr=0.01, opt="adam", loss="cse", mr=0.001)
m.summary()4.2.1 Train model
For the sake of simplicity, I am using only 1000 samples from this test. Additionally, we will use 100 testing samples too.
m.train(x[:1000], y[:1000], epochs=100, batch_size=32, val_x=xt[:100], val_y=yt[:100])The validation accuracy of the model will not be that satisfactory but we can give it a try.
After 70th epoch:
Epoch: 70, Time: 310.139sec
Train Loss: 1707.1975 Train Accuracy: 76.7%
Val Loss: 320.0215 Val Accuracy: 63.0% When using entire datasets, the model's performance will be great.
4.3 Test 2:- Model with 2 Conv2d and Output Layer
m.add(Conv2d(input_shape = (28, 28, 1), filters = 8, padding=None, kernel_size=(3, 3), activation="relu"))
m.add(Conv2d(filters=16, kernel_size=(3, 3), padding=None, activation="relu"))4.3.1 Train model
Let's take 10000 training samples and 500 validation samples. The time to perform an epoch will be huge but the accuracy will be great.
m.train(x[:10000], y[:10000], epochs=100, batch_size=32, val_x=xt[:500], val_y=yt[:500])
Output is something like the below:-
Epoch: 0, Time: 10528.569sec
Train Loss: 21003.3815 Train Accuracy: 53.89%
Val Loss: 1072.7608 Val Accuracy: 54.0%
Epoch: 1, Time: 11990.521sec
Train Loss: 16945.815 Train Accuracy: 67.44%
Val Loss: 845.8146 Val Accuracy: 68.0%
Epoch: 2, Time: 10842.482sec
Train Loss: 14382.4224 Train Accuracy: 72.69%
Val Loss: 790.7897 Val Accuracy: 70.2%
Epoch: 3, Time: 9787.258sec
Train Loss: 10966.7249 Train Accuracy: 80.29%
Val Loss: 585.6976 Val Accuracy: 78.8%
Epoch: 4, Time: 10025.688sec
Train Loss: 9367.4941 Train Accuracy: 83.1%
Val Loss: 487.3858 Val Accuracy: 81.8% It is clear that our model's performance will be good after training more with more data. To be honest, our model's performance is not as good as
kerasbut it is worth trying to code it from scratch.
4.4 Test 3:- A complex model
Let's test our new model, which will have all previously assumed layers.
m = CNN()
m.add(Conv2d(input_shape = (28, 28, 1), filters = 4, padding=None, kernel_size=(3, 3), activation="relu"))
m.add(Pool2d(kernel_size=(2, 2)))
m.add(Conv2d(filters=8, kernel_size=(3, 3), padding=None, activation="relu"))
m.add(Dropout(0.2))
m.add(Flatten())
m.add(FFL(neurons = 10, activation='softmax'))
m.compile_model(lr=0.001, opt="adam", loss="cse")
m.summary()
m.train(x[:5000], y[:5000], epochs=100, batch_size=32, val_x=xt[:500], val_y=yt[:500]) Note that, since this model is huge(has many layers) the time to perform a single epoch might be huge so I am taking only 5000 training examples and 500 testing samples.
The result on my machine is:-
Input Output Shape Activation Bias Parameters
Layer Name
Input Layer (28, 28, 1) (26, 26, 4) relu True 40
Pool2d1 (26, 26, 4) (13, 13, 4) None False 0
Conv2d2 (13, 13, 4) (11, 11, 8) relu True 296
Dropout3 (11, 11, 8) (11, 11, 8) None False 0
Flatten4 (11, 11, 8) 968 None False 0
Out Layer(FFL) 968 10 softmax True 9690
Total Parameters: 10026Total 5000 samples.
Training samples: 5000 Validation samples: 500.
Total 157 batches, most batch has 32 samples.
Epoch: 0:
Time: 1640.885sec
Train Loss: 99970.6308 Train Accuracy: 15.52%
Val Loss: 10490.2164 Val Accuracy: 13.8% The first epoch doesn't seem that much of satisfactory but what might be the other epoch?
Epoch: 10:
Time: 1295.361sec
Train Loss: 37848.7813 Train Accuracy: 57.68%
Val Loss: 4674.9309 Val Accuracy: 53.4%It is quite clear that the model is progressing slowly. And 22nd epoch is:-
Epoch: 22:
Time: 1944.176sec
Train Loss: 22731.3455 Train Accuracy: 76.42%
Val Loss: 3017.2488 Val Accuracy: 69.2%
Epoch: 35:
Time: 1420.809sec
Train Loss: 17295.6898 Train Accuracy: 83.1%
Val Loss: 2358.6877 Val Accuracy: 76.2% A similar model on keras gives 90+ accuracy within the 5th epoch but the good thing about our model is, it is training.
4.5 Test 4:- A complex model
Our model doesn't seem to do great on previous complex architecture. But what if we modified it a little bit? I am using my days to train these models and I have also done lots of hit and trial also.
m = CNN()
m.add(Conv2d(input_shape = (28, 28, 1), filters = 4, padding=None, kernel_size=(3, 3), activation="relu"))
m.add(Conv2d(filters=8, kernel_size=(3, 3), padding=None, activation="relu"))
m.add(Pool2d(kernel_size=(2, 2)))
m.add(Flatten())
m.add(FFL(neurons = 64, activation = "relu"))
m.add(Dropout(0.1))
m.add(FFL(neurons = 10, activation='softmax'))
m.compile_model(lr=0.01, opt="adam", loss="cse")
m.summary()
m.train(x[:10000], y[:10000], epochs=100, batch_size=32, val_x=xt[:500], val_y=yt[:500]) The summary is:-
Input Output Shape Activation Bias Parameters
Layer Name
Input Layer (28, 28, 1) (26, 26, 4) relu True 40
Conv2d1 (26, 26, 4) (24, 24, 8) relu True 296
Pool2d2 (24, 24, 8) (12, 12, 8) None False 0
Flatten3 (12, 12, 8) 1152 None False 0
FFL4 1152 64 relu True 73792
Dropout5 64 64 None False 0
Out Layer(FFL) 64 10 softmax True 650
Total Parameters: 74778Model's Performance is:
Epoch: 5:
Time: 40305.135sec
Train Loss: 1412678.6095 Train Accuracy: 22.43%
Val Loss: 72887.904 Val Accuracy: 24.6%
Epoch: 11:
Time: 7287.762sec
Train Loss: 512155.8547 Train Accuracy: 53.53%
Val Loss: 28439.2441 Val Accuracy: 51.6%
Epoch: 14:
Time: 5984.871sec
Train Loss: 356893.9608 Train Accuracy: 62.85%
Val Loss: 19256.6702 Val Accuracy: 61.0% Model is progressing......
5 Bonus Topics
- Good thing, these topics are interesting.
- Bad thing, you are on your own(but you can leave a comment if explanation needed)
5.1 Save Model
Let's save our model created by Convolutional Neural Networks from Scratch. This method can be placed inside the class that is stacking the layers. Else pass the model object.
def save_model(self, path="model.json"):
"""
path:- where to save a model including the filename
saves Json files on a given path.
"""
dict_model = {"model":str(type(self).__name__)}
to_save = ["name", "isbias", "neurons", "input_shape", "output_shape",
"weights", "biases", "activation", "parameters", "filters",
"kernel_size", "padding", "prob", "stride", "kind"]
for l in self.layers:
current_layer = vars(l)
values = {"type":str(type(l).__name__)}
for key, value in current_layer.items():
if key in to_save:
if key in ["weights", "biases"]:
try:
value = value.tolist()
except:
value = float(value)
if type(value)== np.int32:
value = float(value)
if key == "input_shape" or key == "output_shape":
try:
value = tuple(value)
except:
pass
values[key] = value
dict_model[l.name] = values
json_dict = json.dumps(dict_model)
with open(path, mode="w") as f:
f.write(json_dict)
print("\nModel Saved.")
save_model(m)In the last line of the above code, we are calling a method to save our model. If we looked at our local directory, then there is a JSON file.
5.2 Load Model
This method can be treated as an independent method.
def load_model(path="model.json"):
"""
path:- the path of model file including filename
returns:- a model
"""
models = {"CNN": CNN}
layers = {"FFL": FFL, "Conv2d": Conv2d, "Dropout": Dropout, "Flatten": Flatten, "Pool2d": Pool2d}
with open(path, "r") as f:
dict_model = json.load(f)
model = dict_model["model"]
model = models[model]()
for layer, params in dict_model.items():
if layer != "model":
lyr_type = layers[params["type"]]
if lyr_type == FFL:
lyr.neurons = params["neurons"]
lyr = layers[params["type"]](neurons=params["neurons"])
if lyr_type == Conv2d:
lyr = layers[params["type"]](filters=int(params["filters"]), kernel_size=params["kernel_size"], padding=params["padding"])
lyr.out = np.zeros(params["output_shape"])
params["input_shape"] = tuple(params["input_shape"])
params["output_shape"] = tuple(params["output_shape"])
if lyr_type == Dropout:
lyr = layers[params["type"]](prob=params["prob"])
try:
params["input_shape"] = tuple(params["input_shape"])
params["output_shape"] = tuple(params["output_shape"])
except:
pass
if lyr_type == Pool2d:
lyr = layers[params["type"]](kernel_size = params["kernel_size"], stride=params["stride"], kind=params["kind"])
params["input_shape"] = tuple(params["input_shape"])
try:
params["output_shape"] = tuple(params["output_shape"])
except:
pass
if lyr_type == Flatten:
params["input_shape"] = tuple(params["input_shape"])
lyr = layers[params["type"]](input_shape=params["input_shape"])
lyr.name = layer
lyr.activation = params["activation"]
lyr.isbias = params["isbias"]
lyr.input_shape = params["input_shape"]
lyr.output_shape = params["output_shape"]
lyr.parameters = int(params["parameters"])
if params.get("weights"):
lyr.weights = np.array(params["weights"])
if params.get("biases"):
lyr.biases = np.array(params["biases"])
model.layers.append(lyr)
print("Model Loaded...")
return model
mm = load_model()
mm.summary()
m.predict(x[0]) == mm.predict(x[0])On the above block of code, we tried to load a model. I am not going to describe much here but we are printing a summary and then checking if the prediction from the original model and loaded model is right or wrong. If our model is loaded properly, then the array of all True will be printed.
Upsample Layer
Note that, the Pooling Layer can be called a downsampling layer because it takes samples of pixels and returns a new image with a shape lesser than the original image. And the opposite of this layer is Upsample Layer. Upsample layer generally increases the size of the shape, in more simple words, it zooms the image. And if we see at the configuration of YOLO(You Only Look Once) authors have used multiple times Upsample Layer. In a simpler case, I am doing the pixel expansion.
Let's take an example(in my case):
$$
\begin{pmatrix}
12 & 10 \\
101 & 88 \end{pmatrix}
$$
The output after the kernel (2, 2) will be(the kernel here will not exactly be the kernel like on Maxpool or CNN but it will be used as expansion rate of (row, col)):-
$$
\begin{pmatrix}
12 & 12 & 10 & 10\\
12 & 12 & 10 & 10\\
101 & 101 & 88 & 88\\
101 & 101 & 88 & 88\end{pmatrix}
$$
This is just a simple case of Upsampling, and I have not done much research about it.
class Upsample:
def __init__(self, kernel_size = (2, 2)):
self.input_shape=None
self.output_shape = None
self.input_data= None
self.output = None
self.isbias = False
self.activation = None
self.parameters = 0
self.delta = 0
self.weights = 0
self.bias = 0
self.delta_weights = 0
self.delta_biases = 0
self.kernel_size = kernel_size
self.stride = self.kernel_size
def set_output_shape(self):
shape = self.input_shape
self.output_shape = (shape[0] * self.kernel_size[0], shape[1] * self.kernel_size[1], shape[2])
self.weights = 0
def apply_activation(self, image):
stride = self.stride
kshape = self.kernel_size
self.input_shape = image.shape
self.set_output_shape()
rstep = stride[0]
cstep = stride[1]
self.out = np.zeros(self.output_shape)
shape = self.output_shape
for nc in range(shape[2]):
cimg = []
rv = 0
i = 0
for r in range(kshape[0], shape[0]+1, rstep):
cv = 0
j = 0
for c in range(kshape[1], shape[1]+1, cstep):
self.out[rv:r, cv:c] = image[i, j]
j+=1
cv+=cstep
rv+=rstep
i+=1
return self.out
def backpropagate(self, nx_layer):
"""
Gradients are passed through an index of the largest value.
"""
layer = self
stride = layer.stride
kshape = layer.kernel_size
image = layer.input
shape = image.shape
layer.delta = np.zeros(shape)
cimg = []
rstep = stride[0]
cstep = stride[1]
shape = nx_layer.delta.shape
for f in range(shape[2]):
i = 0
rv = 0
for r in range(kshape[0], shape[0]+1, rstep):
cv = 0
j = 0
for c in range(kshape[1], shape[1]+1, cstep):
dout = nx_layer.delta[rv:r, cv:c, f]
layer.delta[i, j, f] = dout
j+=1
cv+=cstep
rv+=rstep
i+=1 I edited the code of Pool2d for this and backpropagate is a bit different. You can test this code by:-
us = Upsample(kernel_size=(1, 3))
img = us.apply_activation(x_train[0].reshape(28, 28, 1))
plt.imshow(img.reshape(28, 28*3))Visualizing Learned Features
Well, we trained a model but what actually did a model learned? We will be taking the model that we saved earlier. It is loaded on mm. And now we will loop through all layers and the corresponding weights are visualized.
for l in mm.layers:
if type(l).__name__ == "Conv2d":
for f in range(l.filters):
for c in range(l.weights.shape[2]):
plt.imshow(l.weights[:, :, c, f])
plt.title(f"Layer: {l.name} Filter: {f} Channel: {c}")
plt.show()
if type(l).__name__ == "FFL":
plt.imshow(l.weights)
plt.title(l.name)
plt.show()More on Visualization
How will a test image change through the layers? Let's try to find out. When an image gets into any CNN layer, we apply the filters to each channel and sum them. Our feedforward method has granted us a huge application because we can set the input and output of each layer for the current example. And yes that's what we are using.
timg = x[0]
op = mm.predict(x[0])
for l in mm.layers:
print(l.name)
if type(l).__name__ == "Conv2d":
fig = plt.figure(figsize=(30, 30))
cols = l.filters * 2
rows = 1
f = 0
for i in range(0, cols*rows):
fig.add_subplot(rows, cols, i+1)
if i % 2 == 0:
if f < l.filters:
plt.imshow(l.out[:, :, f], cmap="gray")
else:
if f < l.filters:
cimg = l.weights[:, :, 0, f]
plt.imshow(cimg, cmap='gray')
plt.title(f"Layer: {l.name} Filter: {f}")
f+=1
if type(l).__name__ == "Pool2d":
fig = plt.figure(figsize=(30, 30))
cols = l.out.shape[2] * 2
rows = 1
print("Input\n")
for f in range(l.out.shape[2]):
fig.add_subplot(rows, cols, f+1)
plt.imshow(l.input[:, :, f], cmap="gray")
plt.title(f"Layer: {l.name} Filter: {f}")
plt.show()
fig = plt.figure(figsize=(30, 30))
print("Output\n")
for f in range(l.out.shape[2]):
fig.add_subplot(rows, cols, f+1)
plt.imshow(l.out[:, :, f], cmap="gray")
plt.title(f"Layer: {l.name} Filter: {f}")
if type(l).__name__ == "Dropout":
try:
fig = plt.figure(figsize=(30, 30))
cols = l.out.shape[2] * 2
rows = 1
print("Input\n")
for f in range(l.out.shape[2]):
fig.add_subplot(rows, cols, f+1)
plt.imshow(l.input[:, :, f], cmap="gray")
plt.title(f"Layer: {l.name} Filter: {f}")
plt.show()
fig = plt.figure(figsize=(30, 30))
print("Output\n")
for f in range(l.out.shape[2]):
fig.add_subplot(rows, cols, f+1)
plt.imshow(l.out[:, :, f], cmap="gray")
plt.title(f"Layer: {l.name} Filter: {f}")
except:
pass
plt.show()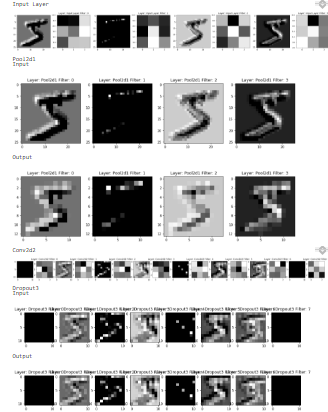
This ends the Convolutional Neural Networks from scratch part of the blog. There are many other algorithms done from scratch and available in this site too.
6 References:¶
I have not done all these codes by myself. I have tried to give credit and references whenever I borrowed concepts and codes. I got help from googling and mostly StackOverflow. However, I have to mention some the great resources at last:-
7 You might like to view:-¶
- Writing Popular Machine Learning Optimizers from Scratch on Python
- Writing Image Processing Class From Scratch on Python
- Writing a Deep Neural Network from Scratch on Python
- Convolutional Neural Networks from Scratch on Python
For the production phase, it is always the best idea to use frameworks but for the learning phase, doing Convolutional Neural Networks from Scratch is a great idea. I also got suggestions from friends that, Prof. Andrew Ng's contents drive us through scratch but I never got a chance to watch one. I am sharing a notebook and repository link also. In the next blog, I will try to do RNN from scratch. Please leave feedback, and if you find this good content then sharing is caring. Thank you for your time and please ping me on[Twitter](https://twitter.com/Quassarianviper). You can find all these files under ML From Basics.